Abstract
In this paper we propose a new framework to compare and classify temporal sequences. The proposed approach captures the underlying dynamics of the data while avoiding expensive estimation procedures, making it suitable to process large numbers of sequences. The main idea is to first embed the sequences into a Riemannian manifold by using positive definite regularized Gram matrices of their Hankelets. The advantages of the this approach are: 1) it allows for using non-Euclidean similarity functions on the Positive Definite matrix manifold, which capture better the underlying geometry than directly comparing the sequences or their Hankel matrices; and 2) Gram matrices inherit desirable properties from the underlying Hankel matrices: their rank measure the complexity of the underlying dynamics, and the order and coefficients of the associated regressive models are invariant to affine transformations and varying initial conditions. The benefits of this approach are illustrated with extensive experiments in 3D action recog- nition using 3D joints sequences. In spite of its simplicity, the performance of this approach is competitive or better than using state-of-art approaches for this problem. Further, these results hold across a variety of metrics, supporting the idea that the improvement stems from the embedding itself, rather than from using one of these metrics.
Problem
The problem is to find an efficient distance function that compares the dynamics of temporal sequences. Then a fast classification or clustering can be achieved.
Proposed Method
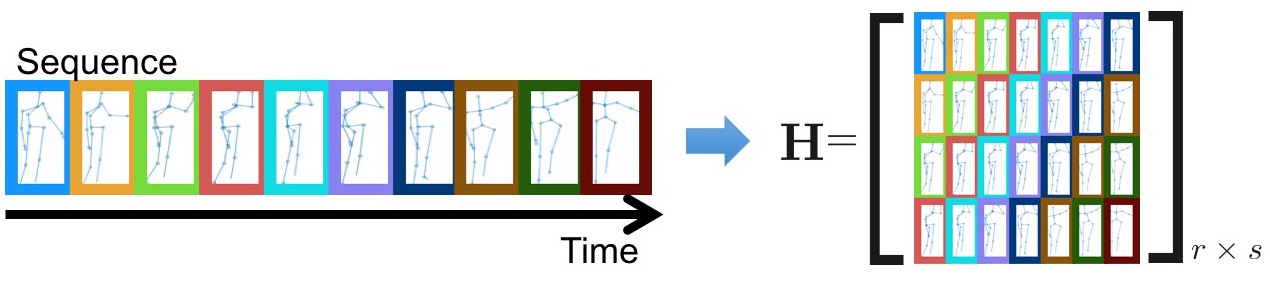

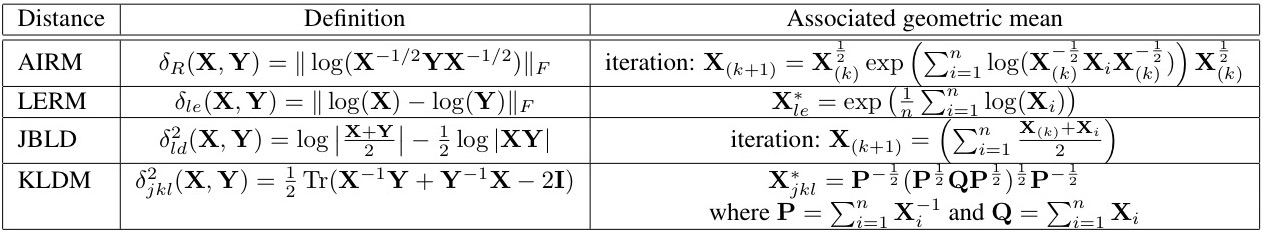
Experiment Results
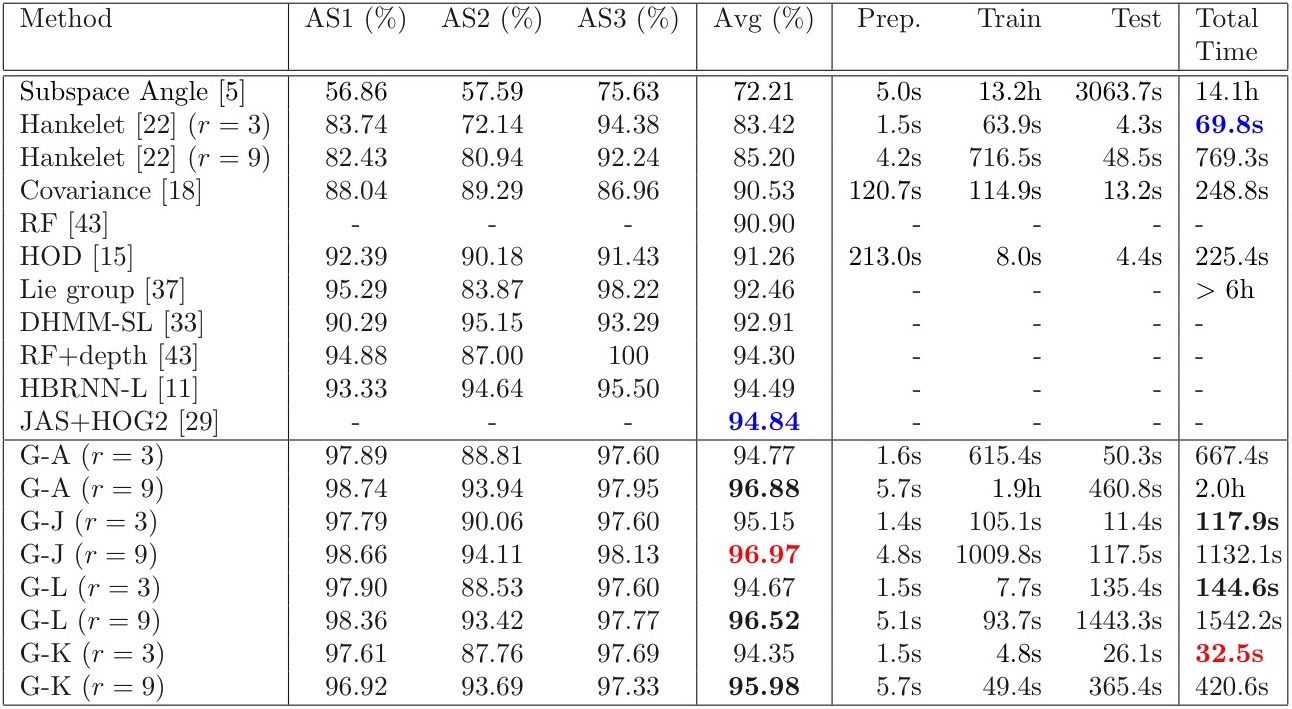
We demonstrated the accuracy and efficiency of our method with MSR3D Action, Berkeley MHAD, UT Kinect and HDM05 datasets. Please find all results in the paper.
Experiment results on MSR3D Action dataset
Publication
Xikang Zhang, Yin Wang, Mengran Gou, Mario Sznaier, and Octavia Camps: Efficient Temporal Sequence Comparison and Classification using Gram Matrix Embeddings On a Riemannian Manifold. CVPR 2016